switchSENSE® technology
The heliX® biosensor instrument enables you to collect comprehensive biophysical data
in one single measurement using the switchSENSE® technology.
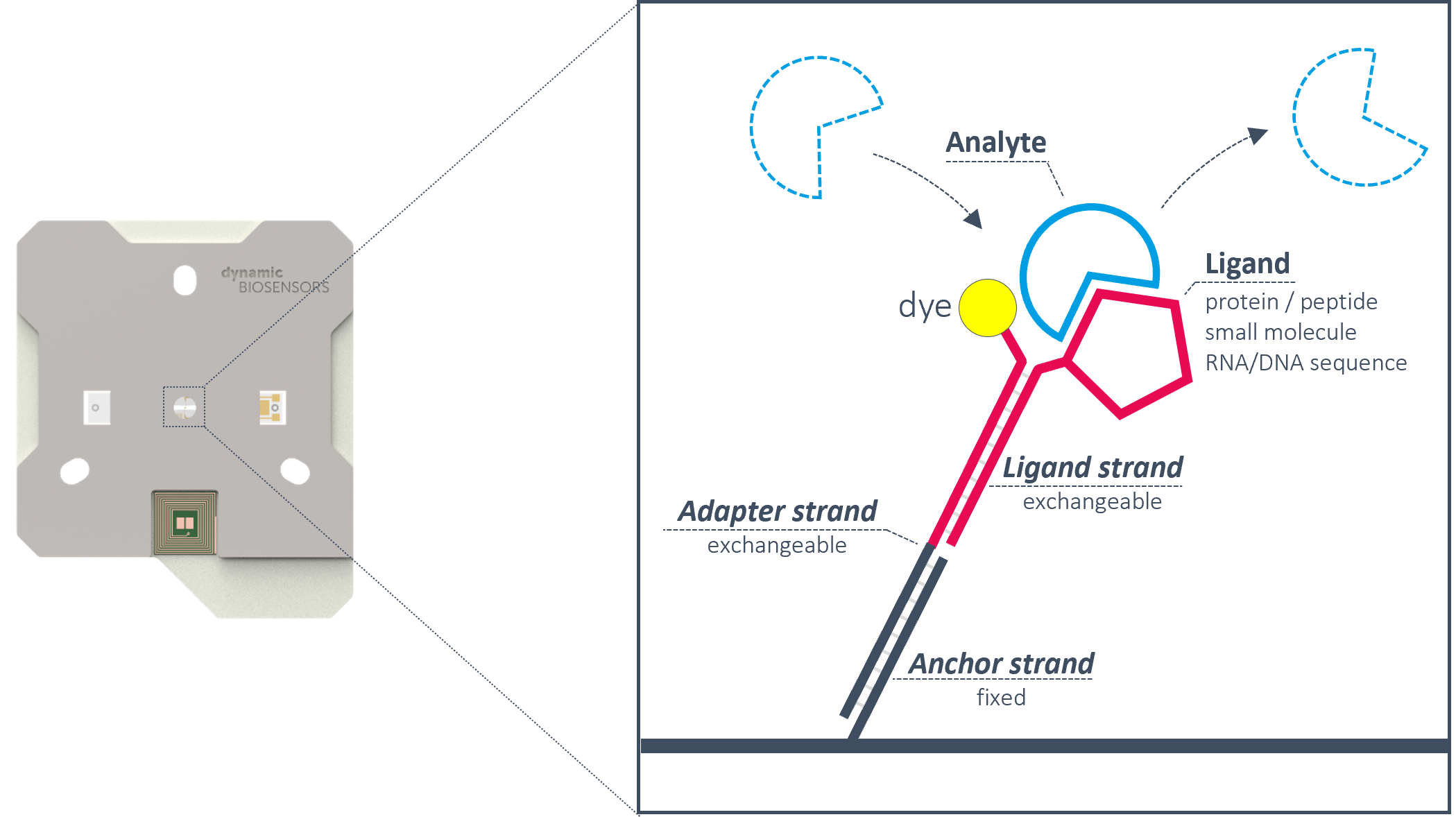
switchSENSE® is based on customizable DNA nanolevers on a chip surface. Different measurement modes allow a multiparameter characterization of biomolecules.
All measurement modes are based on changes in the fluorescent intensity of a dye attached to the nanolever. The fluorescence depends on the local environment of the dye as well as the distance of the dye from the quenching gold electrode surface of the heliX chip.
Measurement Parameters
Comprehensive biophysical information,
in one measurement.
Kinetics (association rate kon, dissociation rate koff)
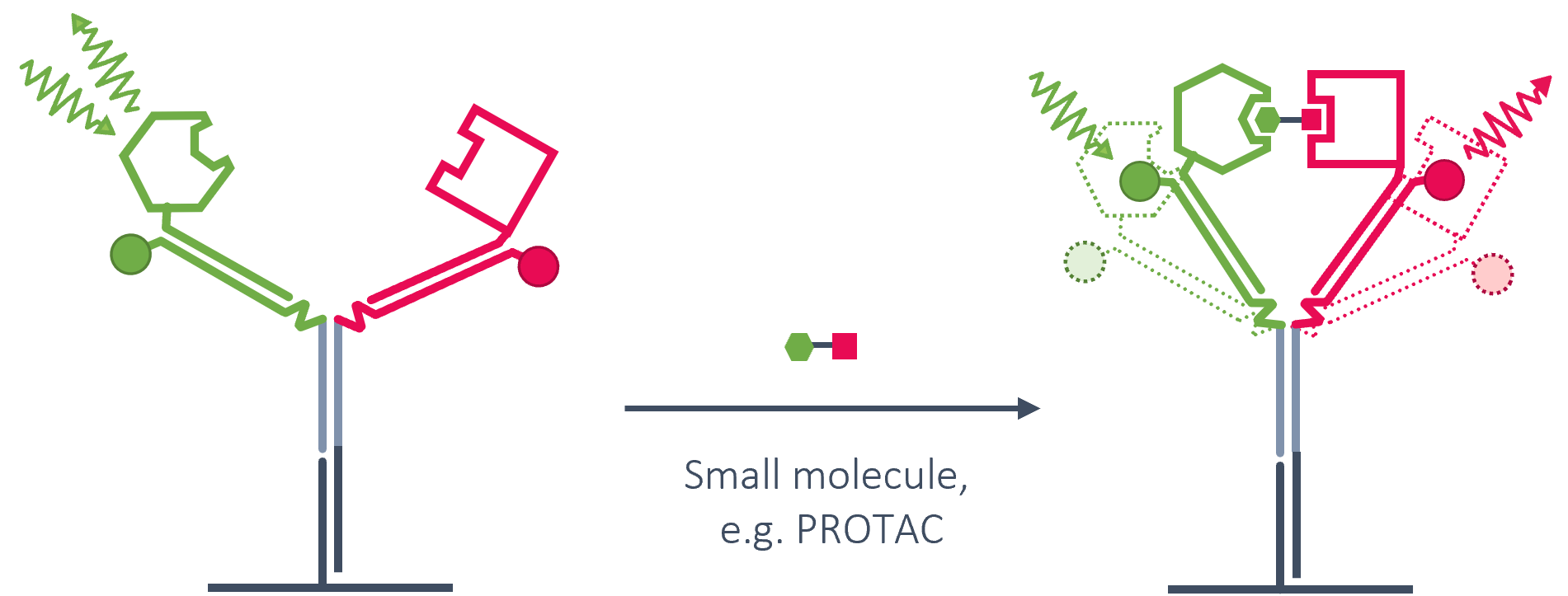
Ternary complex formation / proximity induced binding (e.g. PROTACs)
Affinity, Avidity (Kd)
Conformational Changes
Dose response (EC, IC50)
Relative changes in protein size
Multispecific interactions (e.g. bispecific antibodies)
Nucleic acid enzyme binding and activity (kCAT, KM)
Measurement Modes
The switchSENSE® principle.
THE STATIC MODE
In static mode, the nanolever itself is not moving. The static mode detects the binding of molecules in real-time through changes in the dye’s local environment (fluorescence proximity sensing).
The static measurement mode is also applied when assessing the enzymatic activity of nucleic acid modifying enzymes. These assays exploit the quenching effect of the gold surface.
THE DYNAMIC MODE
In the dynamic measurement mode, the DNA nanolevers are electrically actuated to oscillate at high frequencies.
The measured instantaneous velocity of the nanolever depends on the hydrodynamic friction of the attached molecules. Thus, changes in the upward motion of the nanolever can be used to determine ligand size and shape.
Larger molecules or conformational changes that cause an expansion of the molecule will slow down the movement, whereas smaller molecules or analyte-induced compaction of the molecule will increase the velocity of the motion.
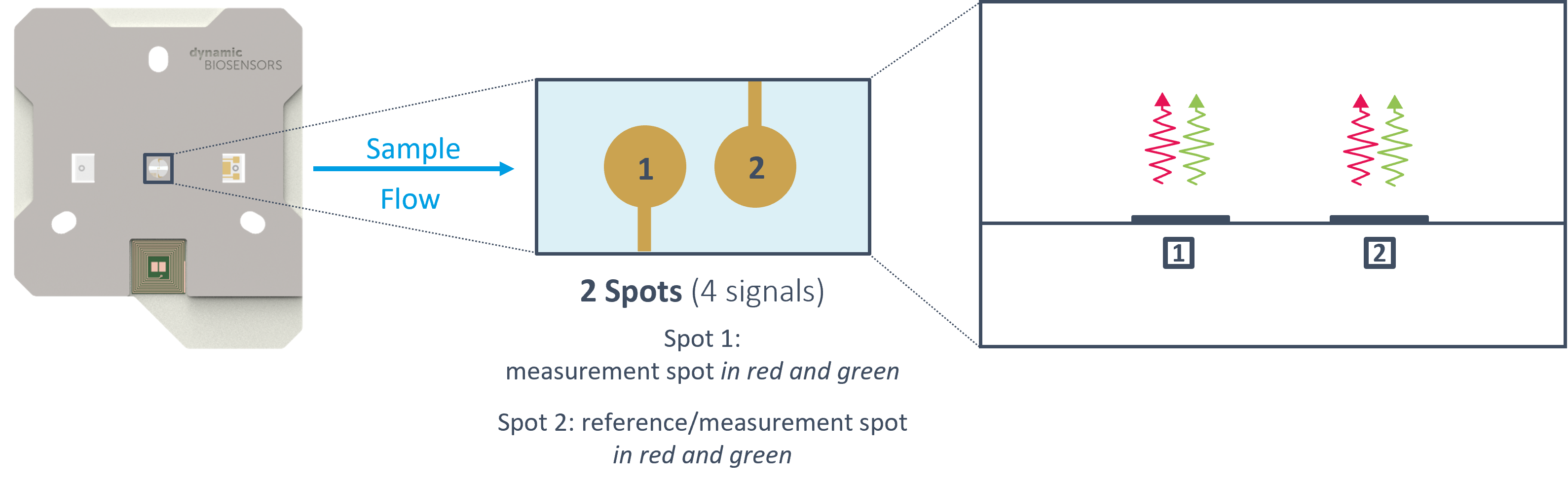
Two-color assays
The unique use of two different fluorophores makes it possible to monitor two independent signals from two interactions
at the same time and on the same sensor spot.
The heliX+ biosensor is equipped with 4 single-photon counters for highest fluorescence sensitivity as well as for the detection of two different fluorophores.
Therefore, two independent signals from two interactions can be monitored at the same time and on the same sensor spot. This allows for many different assay possibilities like multiplexing, bispecific analytes and energy transfer assays (e.g. FRET).
MULTIPLEXING
The heliX chips have two sensor spots. By using the possibility of measuring two independent interactions simultaneously on the same sensor spot, four signals can be monitored in parallel, allowing for real-time referencing of two interactions at the same time or multiplexing of up to four interactions per run.
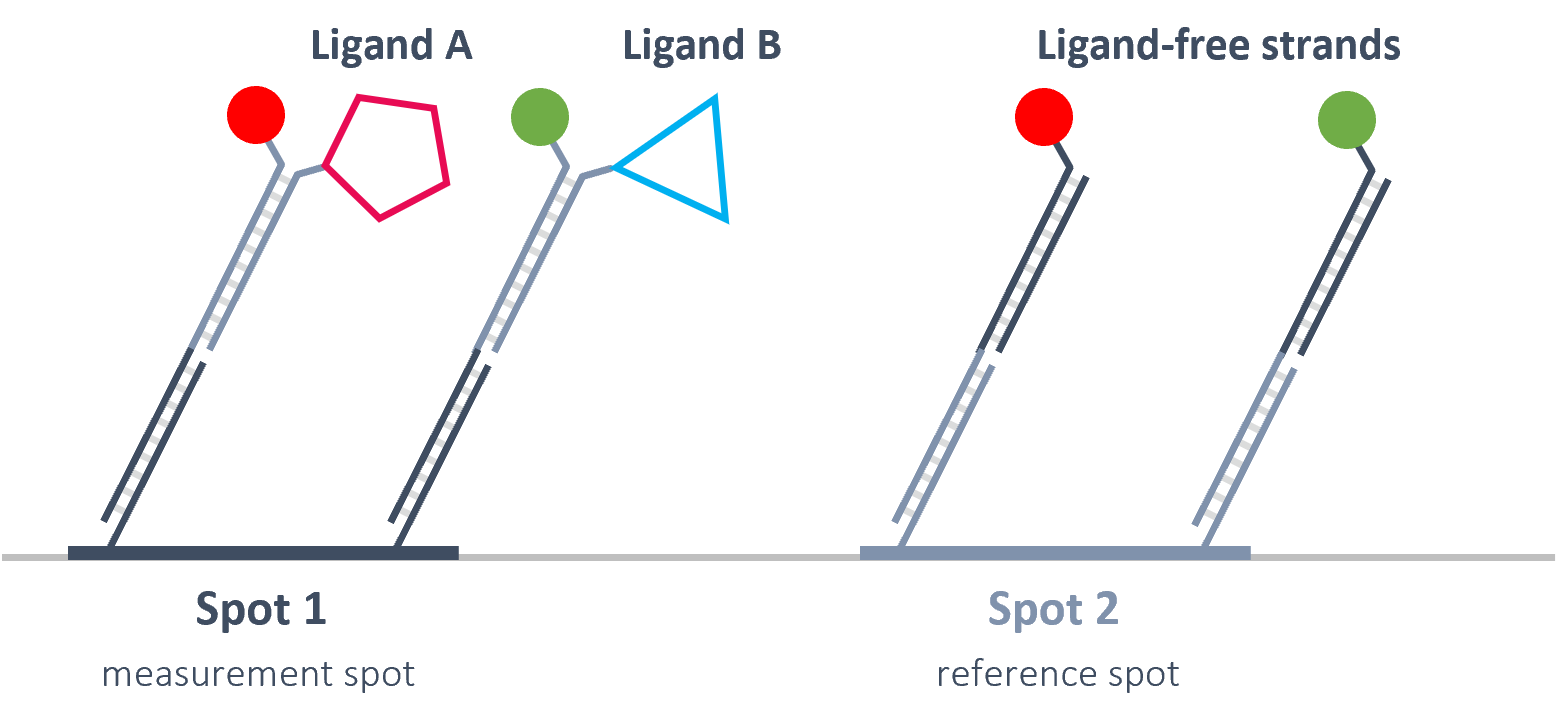
Real-Time referencing of 2 signals.
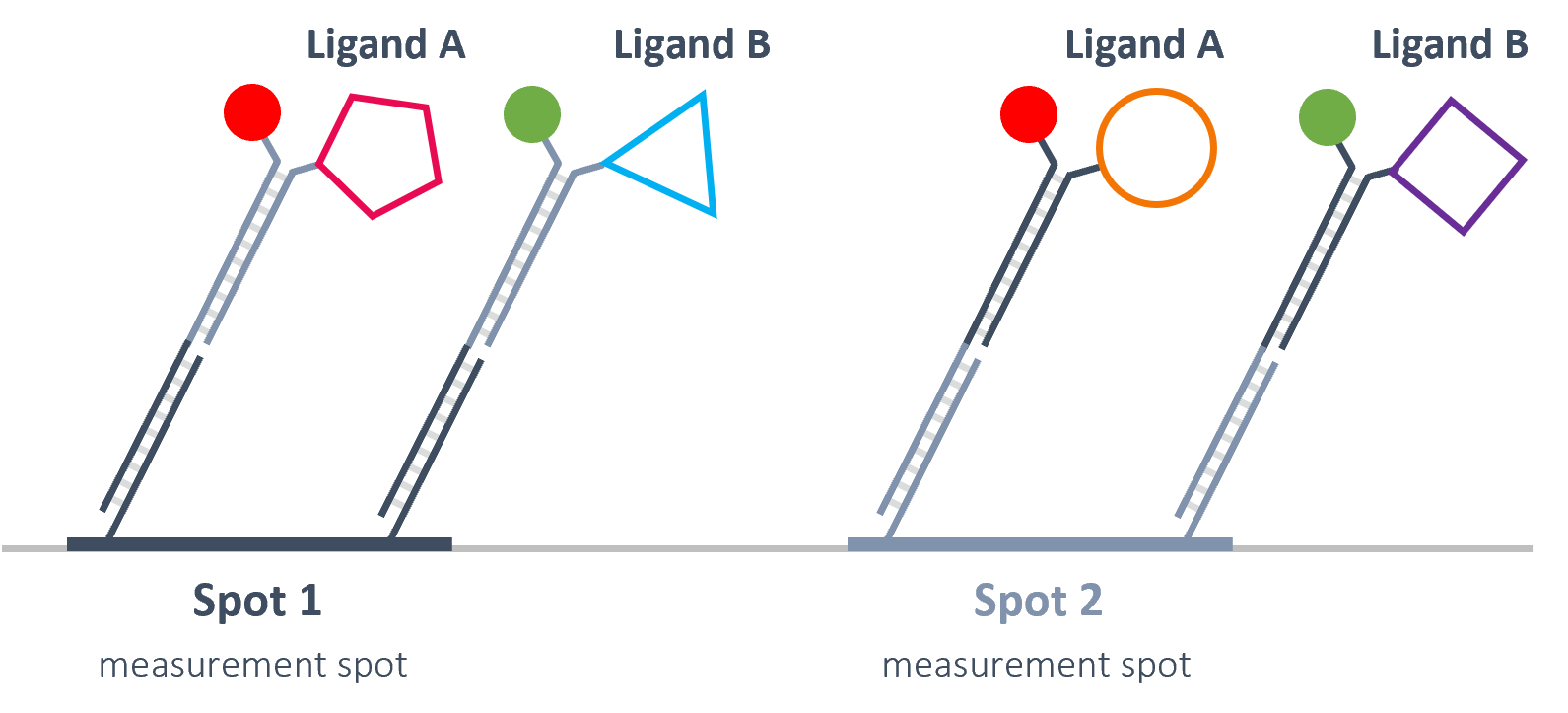
Multiplexing of 4 signals.
BISPECIFIC ANALYTES
One sensor spot can be customized with two different types of nanolevers distinguishable by the different fluorochromes attached to them. That way, the specific binary affinities of a bispecific antibody to its two different antigens can be measured as well as the avidity of the bivalent (ternary) binding.
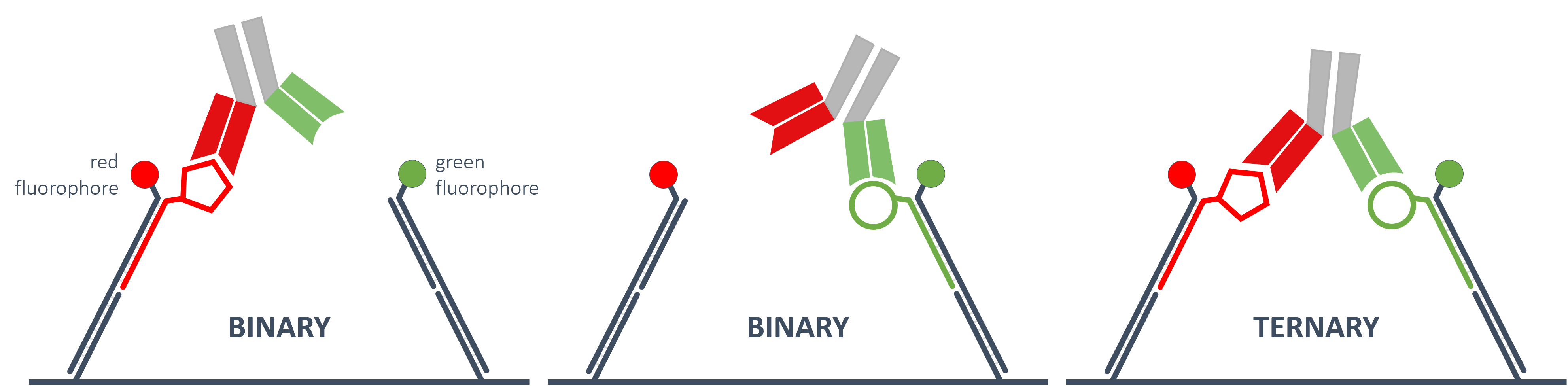
ENERGY TRANSFER ASSAYS
Two color assays additionally offer the possibility of Förster resonance energy transfer (FRET) experiments by using a donor-acceptor fluorochrome pair. This assay format can give unique insights into structural rearrangements of biomolecules, for example RNA secondary structures and loop formation.
DNA Nanotechnology – for unrivaled versatility

We offer a broad portfolio of DNA nanotech-based assay solutions,
which is continuously growing to meet your specific needs.
DNA Y-STRUCTURES FOR CHALLENGING PROTEIN-PROTEIN INTERACTIONS
The portfolio of dual-color assays is further expanded using DNA nanotechnology. Specialized Y-shaped DNA nanostructures bestowed with a fluorochrome FRET-pair are available for the detailed characterization of ternary interactions of bispecific binders. This assay format can be used for example to screen PROTACs and molecular glues or to investigate binding mechanisms of bispecific antibodies.
→ Overcome the challenge of analyzing small molecules that inhibit or stabilize protein-protein interactions [download flyer]
→ Kinetic analysis of ternary and binary binding modes of the bispecific antibody emicizumab [download publication]
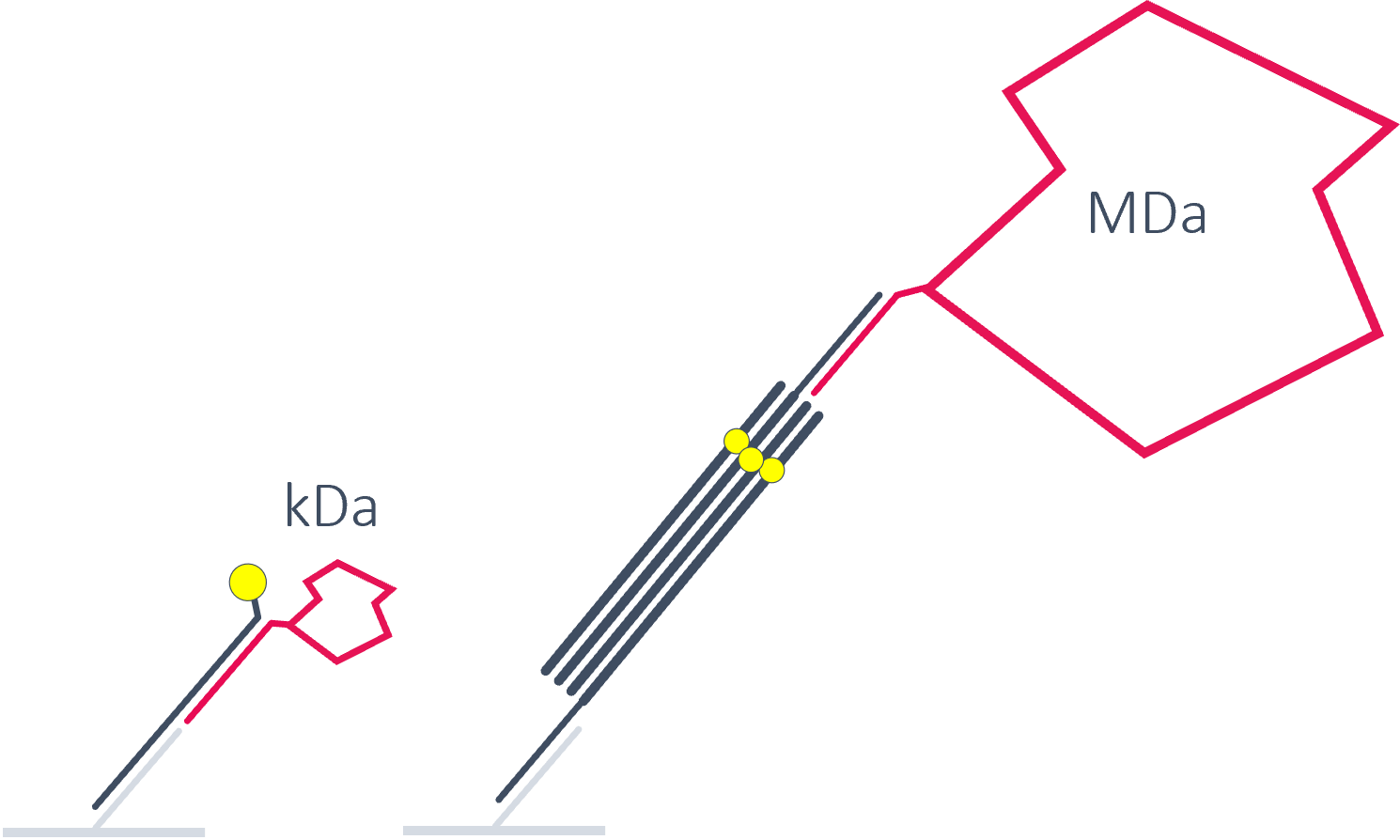
DNA origamis for large proteins and protein complexes
We designed the heliX chip to fit any kind of lever. Short and slender for small proteins, long and rigid for large proteins or protein complexes.
Contact Us
Discuss your specific needs with our specialists.