Brochure I May 2, 2017
Advanced biophysical analysis of mono and bispecific antibody formats with the switchSENSE® Biosensor
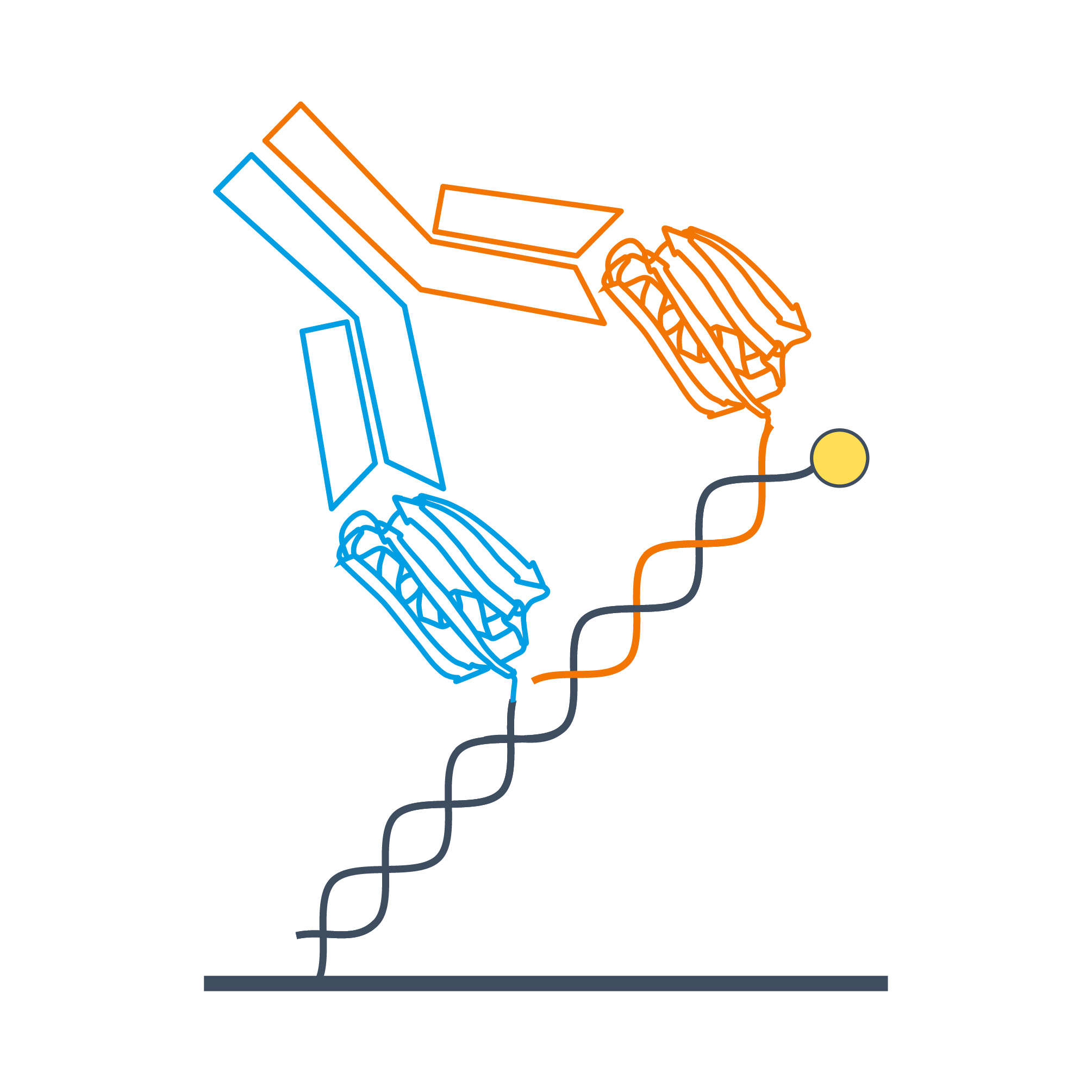
High-affinity and bispecific antibody formats are challenging analytes for interaction analysis systems, because the apparent binding kinetics crucially depend on how the target molecules are presented on the sensor surface. In order to emulate the presentation of heterogeneous antigens on a cell surface with a biosensor platform it is necessary to functionalize the sensor with different antigens at a defined stoichiometry and to control the spatial arrangement of these antigens relative to each other, which has not been feasible up to now. We introduce the DNA-templated assembly of different ligands on a switchSENSE sensor and demonstrate the quantitative measurement of binding cooperativity, i.e., avidity effects. The influence of different ligand arrangements on the binding kinetics, in particular the off-rate, is discussed for antibody formats and bifunctional small proteins. We show how the ligand-to-ligand distance can be controlled with sub-nanometer precision using bifunctional DNA scaffolds, i.e. nanolevers with adjustable arm-lengths. Measurements with these templated ligand arrangements are compared to the conventional case of random ligand adsorption at different densities and steric influences are exemplified. We investigate bispecific interactions with similar (koff,1~koff,2) and dissimilar (koff,1>10 x koff,2) dissociation rates and describe the resulting avidities. Moreover, a comparison of the obtained results to the predictions of a theoretical dissociation model based on monovalent off-rates reveals substantial differences, which underlines the importance of experimental studies for the understanding of binding cooperativity. We believe the introduced workflows will be highly instrumental in the discovery and selection of bispecific biotherapeutics.