For biophysical experiments, a switchSENSE® measurement combines two aspects:
- A fluorescence measurement, where the fluorophore emission intensity is subject to two influences:
- a non-radiative energy transfer to the metal electrode, which depends on the distance between the fluorophore and the metal surface
- quenching or enhancement of the fluorescence intensity of the dye due to the proximity between the dye and molecules bound within its probing environment.
- The electrical control over the orientation of end-tethered DNA nanolevers:
- either by applying a constant potential (mild negative potentials) to the electrode to align the DNA in a fixed, well-defined orientation (usually upright).
- or by applying alternating repulsive and attractive potentials to facilitate a dynamic oscillation of the DNA nanolever orientation.
This implies that a switchSENSE® experiment can be performed using two different approaches: with switching of the DNA nanolevers (dynamic mode) or without switching (static mode). These two measurement modes differ mainly in the electric potentials that are applied to the sensor electrodes. Experiments in dynamic mode are performed under constant orientation switching of the DNA nanolevers, actuated by oscillating potentials, which either attract or repel the DNA backbones.
In contrast to this, experiments in static mode are performed with a constant potential (usually negative) that keeps the DNA nanolevers static at a fixed angle. Figure 2 illustrates the two measurements modes.
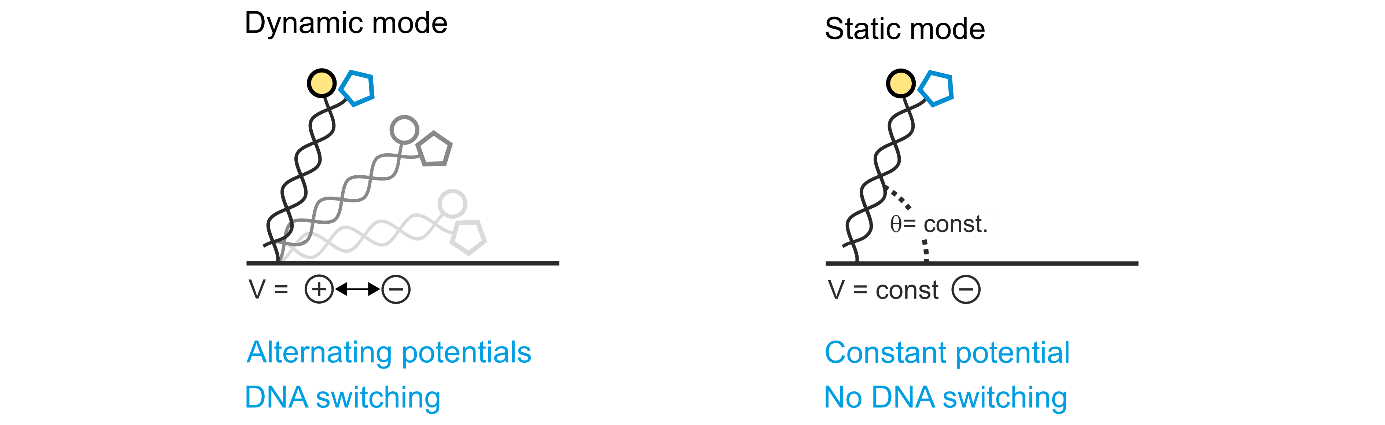
Figure 2: switchSENSE® measurement modes. In dynamic mode, the potentials applied to the sensor electrodes are alternated, which drives the DNA nanolevers to oscillate on the electrode’s surface due to their negatively charged backbone. When using the static mode, usually a constant negative potential is applied, which positions the DNA nanolevers constantly in an upright conformation.
Experiments in dynamic mode provide the following evaluation parameters:
- Molecular dynamics: hydrodynamic changes can be detected as changes in the Dynamic Response.
- Protein sizing: the hydrodynamic diameter (DH) of proteins, protein complexes or of folded nucleic acids can be determined using the Lollipop model.
- Fluorescence proximity sensing (FPS): changes in the local environment of the fluorophore are detected from the changes in the intensity of the fluorescence emission.
- Molecular ruler: changes in the average position of the fluorophore with respect to the gold surface can be determined by observation of the fluorescence intensity.
When to use best:
- When working at salt concentrations up to physiological conditions.
- Experiments with small ligand molecule relative to a larger analyte molecule provide best signal to noise ratio in molecular dynamics.
Advantages compared to static mode:
- Full experimental information content is provided
- Excellent long-term signal stability during kinetic experiments
Experiments in static mode provide the following evaluation parameters:
- Fluorescence proximity sensing (FPS): changes in the local environment of the fluorophore are detected by changes in the intensity of the fluorescence emission.
- Molecular ruler: changes in the average position of the fluorophore with respect to the gold surface can be determined by observation of the fluorescence intensity.
Advantages compared to dynamic mode:
- Improved signal to noise ratio and higher sensitivity for FPS analysis
- Extended chip lifetime