Chip regeneration implies that all ligand molecules immobilized on switchSENSE® electrodes in one flow channel and all potentially bound analyte molecules are completely washed off the sensor surfaces by treatment with a regeneration solution. This leaves single-stranded DNA nanolevers on the sensor electrodes, which can be re-functionalized with fresh ligand molecules by hybridization to the corresponding complementary DNA nanolevers.
The regeneration process contains two crucial steps: the denaturation of the double-stranded DNA nanolevers, leaving single-stranded DNA molecules on the electrodes and successively, the hybridization during which double-stranded nanolevers are formed again. The hybridization of the DNA nanolevers is usually monitored during a switchSENSE® experiment. Figure 10 shows typical fluorescence intensity traces corresponding to the transition of single stranded DNA nanolevers to double stranded DNA nanolevers. The basic effect that allows to observe DNA hybridization on a switchSENSE® sensor electrode is that single-stranded DNA nanolevers and double-stranded DNA nanolevers exhibit remarkably different switching properties. Due to its high flexibility, single stranded DNA can only be switched within the area above the electrode that is subjected to electric field, which extends to only a few nanometers into the electrolyte solution. In contrast to this, double stranded DNA is very rigid, with the consequence that during the repulsive switching phase, parts of the DNA nanolever that are exposed to the electric field push the major part of the nanolever far beyond the electrically affected areas. As stated previously under the Molecular Ruler principle, this translates to an increase in repulsive fluorescence intensity (Fup) and in switching amplitude, which is defined as the difference between the repulsive and attractive fluorescence intensity (ΔF).
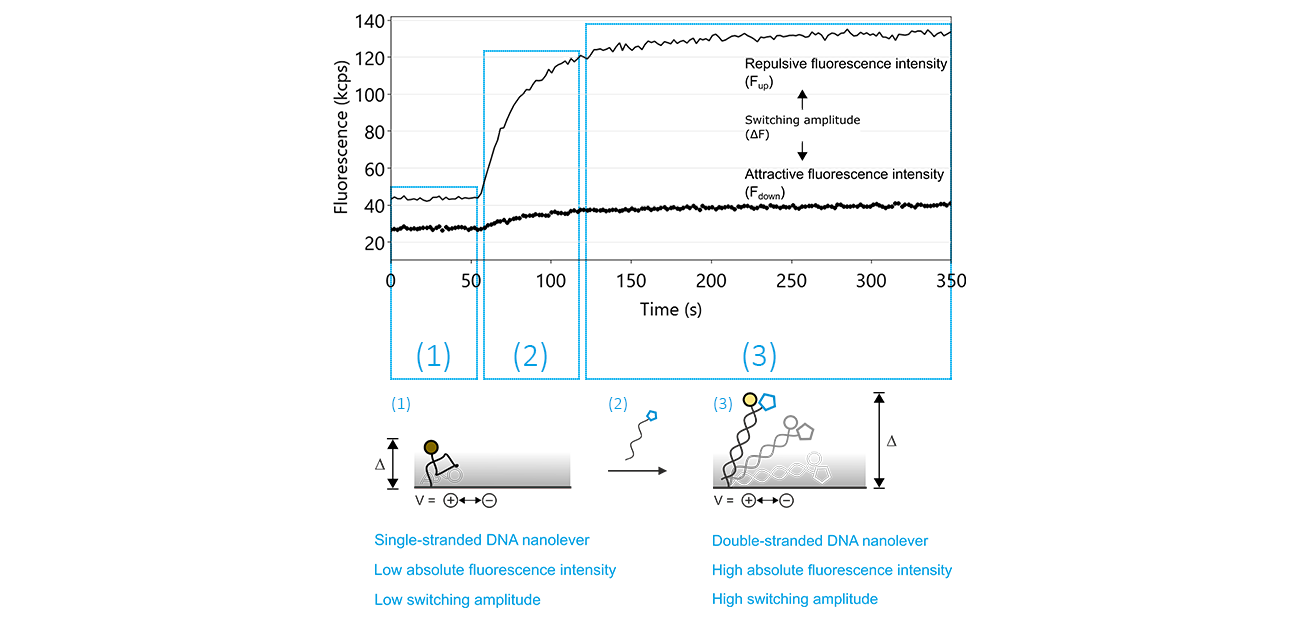
Figure 10: The denaturation of double-stranded DNA nanolevers using a regeneration solution with a basic pH value followed by re-hybridization, yielding double-stranded DNA nanolevers, are the two key features that facilitate the unique regeneration process of switchSENSE® electrodes. After denaturation, which is not recorded by the sensor system, the switching process can be monitored by observing the attractive (Fdown; dotted line) and repulsive (Fup; solid line) fluorescence intensity values. As long as the DNA nanolevers remain in the single-stranded state, the Fup is comparably low, with a reduced switching amplitude (ΔF). The hybridization of the complementary DNA is marked by a significant increase of Fup and therefore of ΔF.
Denaturation of the DNA nanolevers:
The denaturation of the double-stranded DNA nanolevers is automatically carried out by the DRX instrument by treatment of the sensor surfaces with the basic Regeneration Solution, leaving single-stranded DNA on the sensor electrodes. In case this step is not successful, the DNA nanolevers on the sensor electrodes remain double stranded, which prevents hybridization of fresh DNA-ligand conjugates. Figure 11 shows an exemplary data set of a switchSENSE® hybridization routine, during which the denaturation step was not successful. The reason for absence of denaturation or incomplete denaturation is usually shortage of regeneration solution or inactive regeneration solution. The latter is sometimes observed when the regeneration solution was stored in contact with air for an extended timescale.
How do I recognize insufficient denaturation?
- Check the absolute fluorescence intensity at the beginning of the regeneration trace. On successful denaturation, the fluorescence intensity is comparably low (Fup ~ 10 – 50 kcps). If the baseline of the hybridization trace (up to about 50 s after start of the measurement) is significantly higher, this denotes that the DNA nanolevers are still double-stranded.
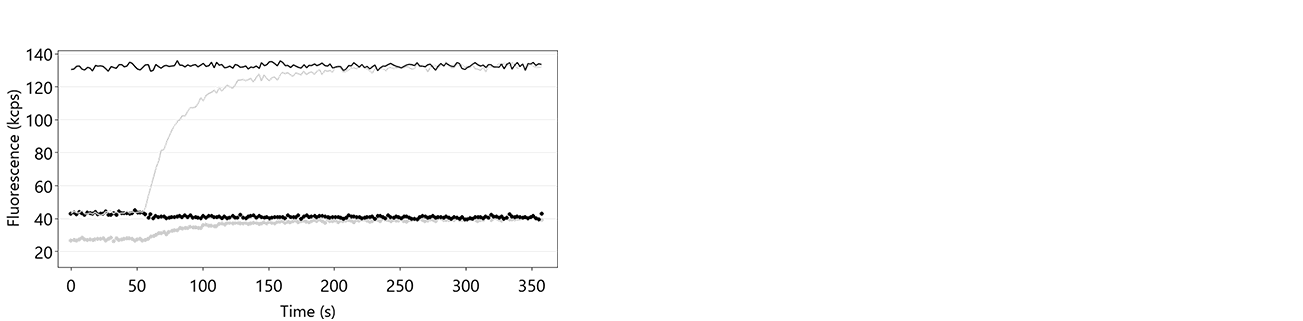
Figure 11: Fup (solid line) and Fdown (dotted line) traces of a hybridization routine after insufficient denaturation of the DNA nanolevers. It is clearly visible that Fup constantly remains at a high intensity level during the entire process, which indicates that there is no transition from single-stranded to double-stranded DNA nanolevers. In grey, the respective hybridization traces on successful denaturation are shown for comparison.
Hybridization of the DNA nanolevers:
After denaturation of the DNA nanolevers, the single-stranded DNA molecules on the sensor electrodes are re-hybridized to double-stranded DNA nanolevers by incubation with the respective complementary DNA molecules. This is a crucial step for switchSENSE® experiments, as it facilitates the immobilization of ligand molecules. Given that the denaturation step was successful, problems with the hybridization step are usually either caused by shortage of DNA solution or by incubation with non-complementary DNA molecules. Furthermore, the use of an insufficient concentration of complementary DNA will result in partial DNA nanolever hybridization. Figure 12 shows fluorescence intensity traces of three independent regeneration routines, which exhibit no or partial DNA nanolever hybridization.
How do I recognize insufficient hybridization?
- Does the fluorescence intensity increase during the hybridization? The hybridization of the complementary DNA is accompanied by a significant increase of the repulsive fluorescence intensity (Fup), caused by the transition of the immobilized DNA molecules from single-stranded to double-stranded state. On successful hybridization with bare complementary DNA, the Fup intensity should at least increase by 100 %.
- Is there a switching amplitude, i.e. is there a significant difference between Fup and Fdown? Even in single-stranded state, switching DNA nanolevers exhibit ΔF values of about 10-20 kcps. If there is no significant difference between Fup and Fdown, this points to the presence of air in the flow channel, which is usually the consequence of an empty autosampler vial.
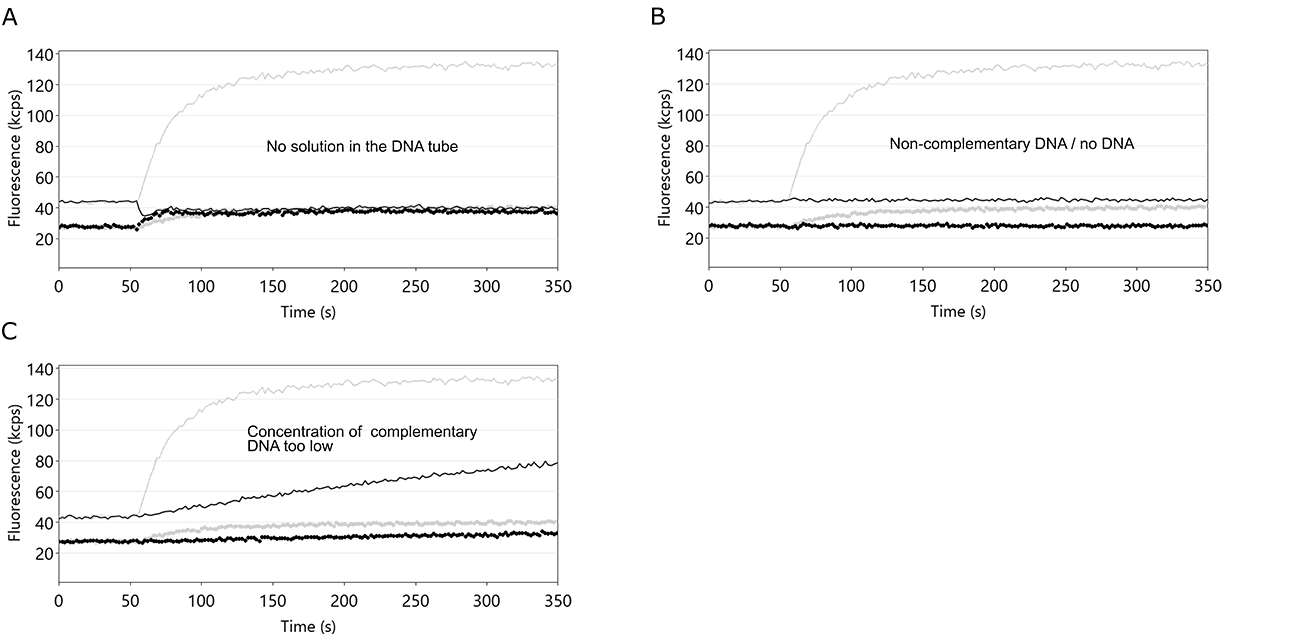
Figure 12: Fluorescence intensity profiles of switchSENSE® hybridization routines. In each example, the hybridization of the DNA nanolevers was hindered by the stated cause. A: an empty DNA vial. If the volume of the complementary DNA solution is not sufficient, air is injected into the flow channel, which immediately ceases the DNA switching motion. Fup and Fdown now show identical values. B: use of non-complementary DNA or no DNA. In both cases, the single-stranded DNA nanolevers on the switchSENSE electrodes cannot hybridize to their complementary DNA and thus remain single-stranded. In this case, Fup and Fdown remain constant. C: concentration of the applied DNA solution is too low. Corresponding to other association kinetics, the hybridization of DNA nanolevers is affected by the concentration of complementary DNA. If the concentration of complementary DNA is too low, the hybridization process is not completed within the time-scale chosen by the switchBUILD software and neither Fup, nor ΔF reach a constant final level. Grey lines depict the case of ideal DNA nanolever hybridization.
What can I do to improve the hybridization efficiency?
Typically, the concentrations of complementary DNA solutions and the corresponding hybridization times suggested by the switchBUILD software are optimized to ensure surface saturation by hybridization. Yet, in some cases, ligand molecules conjugated to the complementary DNA strand can interfere with the hybridization and thus reduce the hybridization speed. In these situations, the hybridization efficiency can be increased with the following measures:
- Increase the concentration of complementary DNA. This is a simple but efficient way to increase the hybridization speed, which scales linearly with the concentration of complementary DNA.
- Increase the hybridization temperature. Elevated temperatures generally accelerate the hybridization velocity as is the case for most biochemical reactions. Secondly, nucleic acid ligands (overhangs) that are attached to the complementary DNA can form secondary structures that hinder the formation of the double-stranded DNA nanolevers. These secondary structures can often be de-stabilized by working at higher temperatures. To perform your hybridizations at temperatures above 25 °C, activate the expert mode of switchBUILD. The desired hybridization temperature can then be adjusted in the respective experimental element. Please ensure that the ligand molecules of interest are stable at the chosen elevated hybridization temperature.
- Reduce the salt concentration of DNA conjugate solution: Similar to increased hybridization temperatures, also a reduction of salt concentration in the hybridization solution simultaneously accelerates the general DNA hybridization process and also destabilizes secondary structures of nucleic acid ligands. Please make sure to choose conditions that correspond to at least 10 mM of total monovalent salt concentration to ensure sufficient shielding of the negatively charged DNA backbone.